KI - Image Processing server
## Pablo LasoI am a data scientist and machine learning engineer in Nashville, TN mostly using NLP and machine learning to solve business problems and deliver efficient, scalable solutions.Introduction
In the radiology department of a hospital, there are numerous tedious and time-consuming tasks that doctors or researchers must undertake after an MRI scan. For example, researchers studying Multiple Sclerosis (MS) must use software like the Lesion Segmentation Tool (LST) to segment T2 hyperintense lesions in FLAIR images. Implementing a system that can automatically perform all required image processing steps in a streamlined manner will significantly reduce time and improve overall efficiency.
Server
In this project for the Radiology Department (Neuro MRI research group) at the Nobel Prize-awarding institution, Karolinska Institutet, a server was developed to automate various Image Processing tasks. The server can be run on a properly configured workstation and allows the user to either perform steps manually or automatically. In manual mode, the user can specify which software to use for data processing. In automatic mode, the server will recognize the type of data from multiple studies and choose the appropriate software accordingly. Additionally, the server will identify identical patients based on their Study ID (as they are already anonymized) to facilitate longitudinal studies.
The user interface for the server is shown in the example where different steps can be performed on the right-hand side. The user can choose to run all steps automatically to save time by selecting "AUTO (all steps)". To start the process, the user simply needs to provide the path to the input folder on the local machine and the server will perform anonymization, BIDSyfication (formatting to BIDS standard), and the selected step on each subject. It will also check for possible longitudinal studies in each subject.
Pipeline design
The illustration below shows the simplified workflow of the pipeline, from data export to image processing. Firstly, all DICOM files are exported to a hardware device. Then, the locally running server is responsible for anonymizing, unpacking, converting to NIfTI format, and setting the data in BIDS format. Finally, based on the user's specific request (or the type of images available), various software programs are executed to process the images.
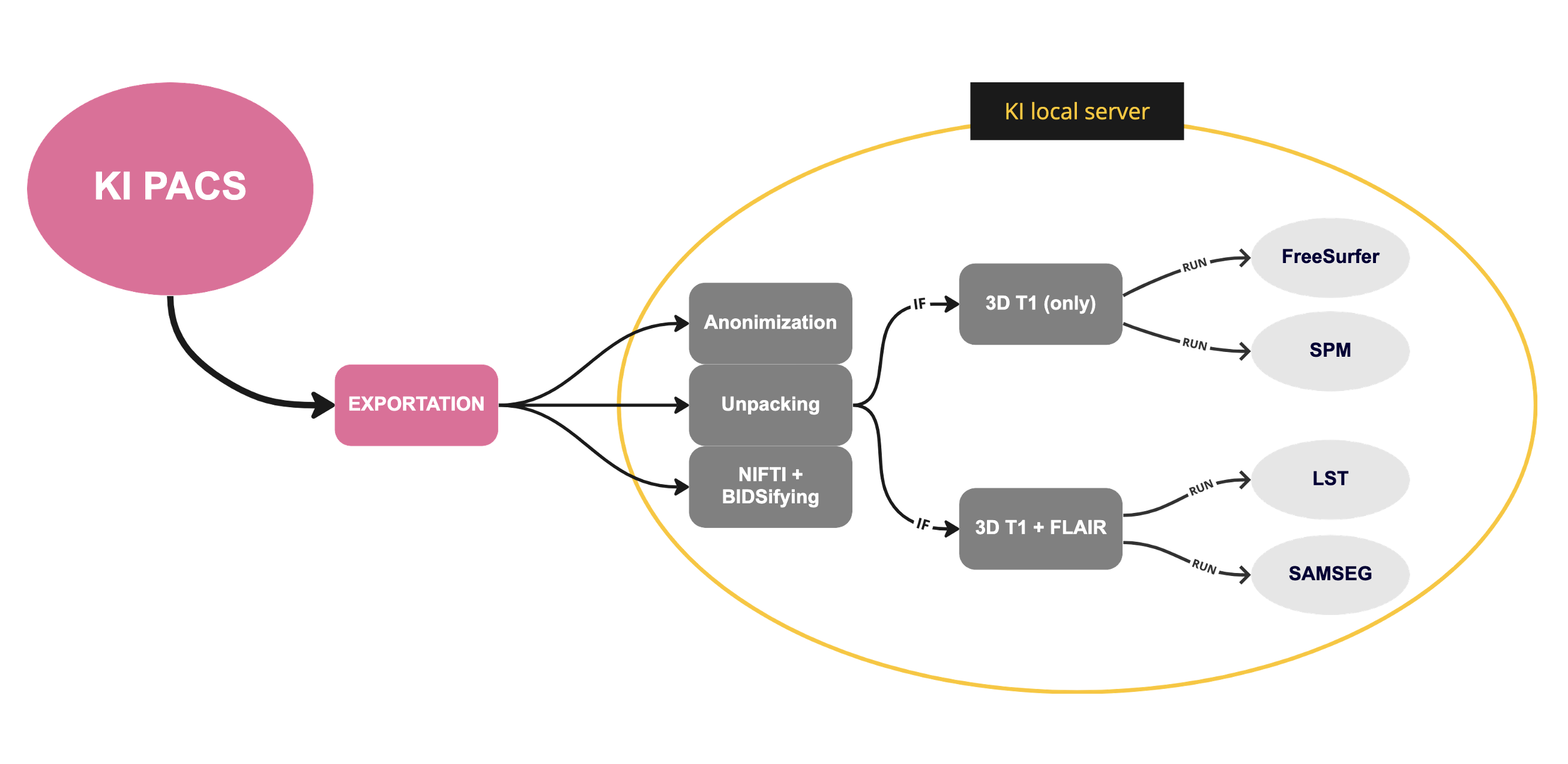
Currently supported programs
- Data preparation (after PACS exportation):
- Anonymization (dicomanon): a MATLAB anonymization function to remove all sensitive data from the DICOM headers and residual files after PACS exportation.
- Unpacking: reading the DICOM metadata to sort images into study protocols, e.g., T1, T2, FLAIR.
- NIFTI converter (dcm2niix): converting DICOM images (.dcm) into NIfTI format (.nii).
- BIDSifying: sorting NIfTIs according to BIDS format, e.g., dwi, anat, func, etc.
- Statistical Parametric Mapping (SPM): an fMRI analysis software package that is run in Matlab. In addition to fMRI analysis, SPM contains toolboxes for performing volume-based morphometry and effective connectivity.
- FreeSurfer: An open source neuroimaging toolkit for processing, analyzing, and visualizing human brain MR images.
- SAMSEG: Sequence Adaptive Multimodal SEGmentation (SAMSEG) is a tool to robustly segment dozens of brain structures from head MRI scans without preprocessing.
- Lesion Segmentation Tool (LST): The toolbox LST is an open source toolbox for SPM that is able to segment T2 hyperintense lesions in FLAIR images, originally developed for the segmentation of MS lesions.
- FSL: a comprehensive library of analysis tools for FMRI, MRI and DTI brain imaging data.
- Spinal Cord Toolbox (SCT): a comprehensive, free and open-source set of command-line tools dedicated to the processing and analysis of spinal cord MRI data.
- miscellaneous: Additional handy tools are added such as a zipper function to compress all data in a given folder in a individual fashion, i.e., subject by subject.